We use biochemical, biophysical and structural methods to define the key macromolecular interactions that regulate important biological processes. Our major focus is the innate immunity pathway for defense against viral infection.
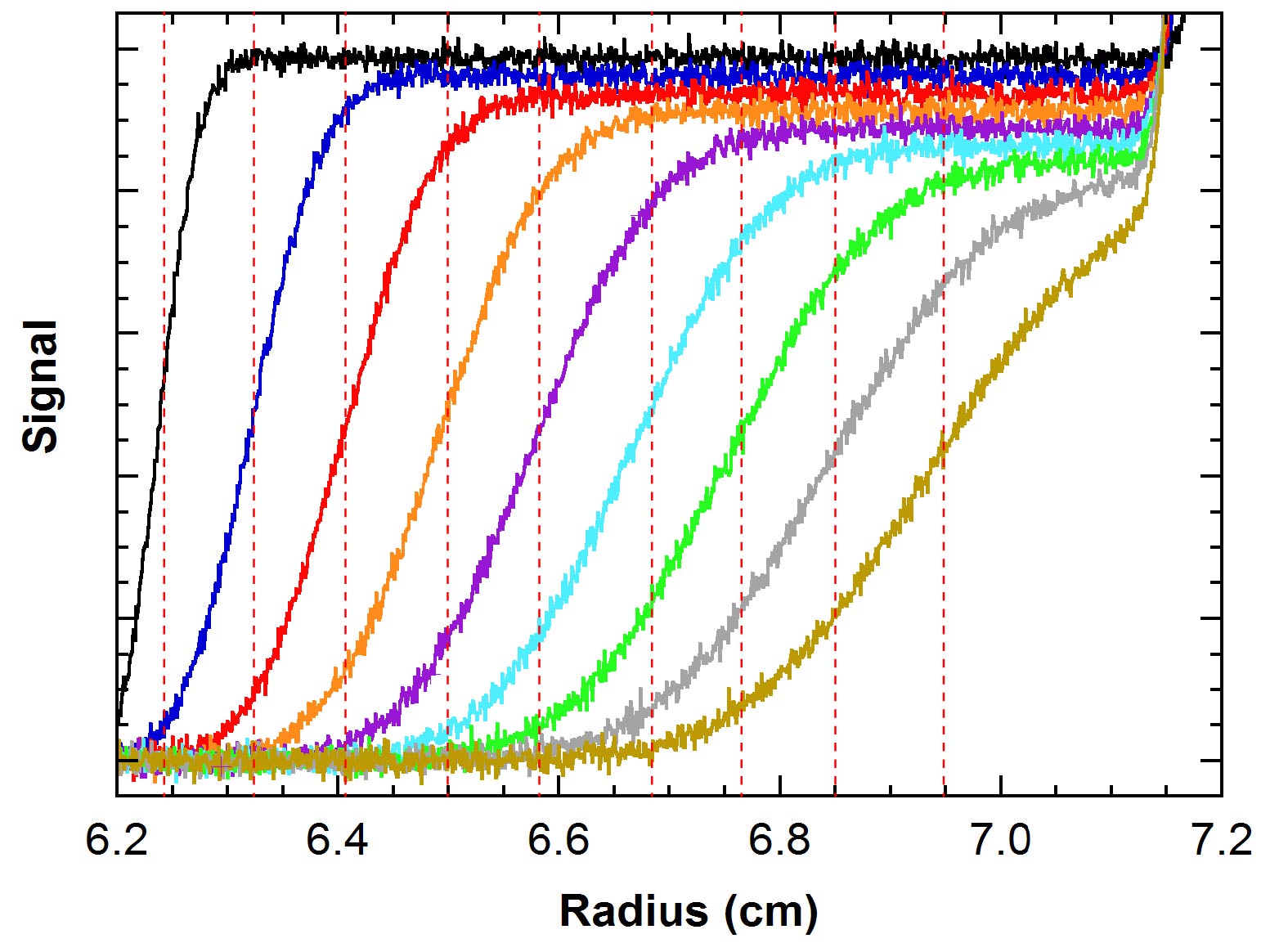
PKR
The RNA activated kinase PKR is a key component of the innate immunity response to viral infection. PKR is induced by interferon that is produced during viral infection and is activated upon binding to dsRNA. Activated PKR blocks protein synthesis within infected cells by phosphorylating the translational initiation factor eIF2α. Our goal is define the molecular mechanism for activation of PKR by RNA.
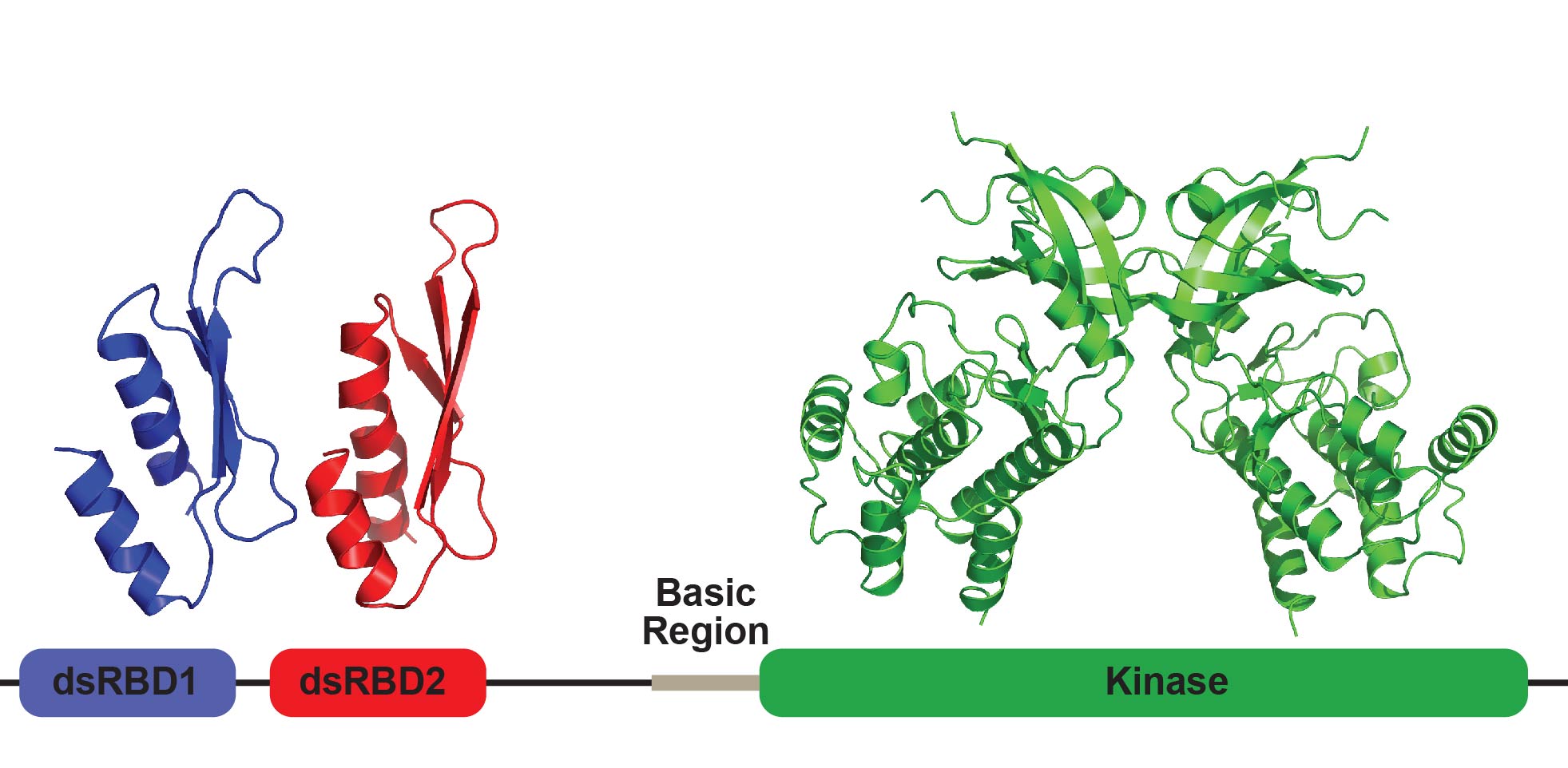
COVID-19 Drug Discovery
COVID-19 is caused by the novel coronavirus SARS-CoV-2. We lack a vaccine and there is an urgent need for antiviral therapies. The viral endoribonuclease EndoU enables coronaviruses to evade the host antiviral defenses by degrading RNAs that would otherwise activate dsRNA sensors such as PKR and MDA5. EndoU activity is required for replication of coronaviruses. In this project we are identifying inhibitors of EndoU and evaluating their antiviral effects against SARS-CoV-2 and other coronaviruses as well as defining how EndoU activity is regulated.
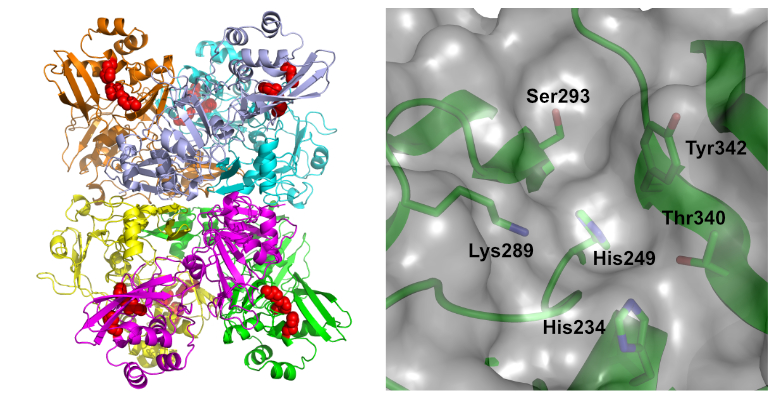
RIG-I-like Receptors
The innate immunity pathway is activated by recognition of conserved molecules that indicate the presence of a pathogen. Cytosolic viral RNAs are detected by a family of RIG-I-like receptors: RIG-I, MDA5, and LGP2. RIG-I recognizes short viral dsRNAs that contain a 5'-triphosphate whereas MDA5 preferentially interacts with longer dsRNA ligands. LGP2 lacks the CARD signaling domains and modulates MDA5 signaling. We are interested in defining the mechanism for assembly of the active signaling complexes of RIG-I and MDA5.
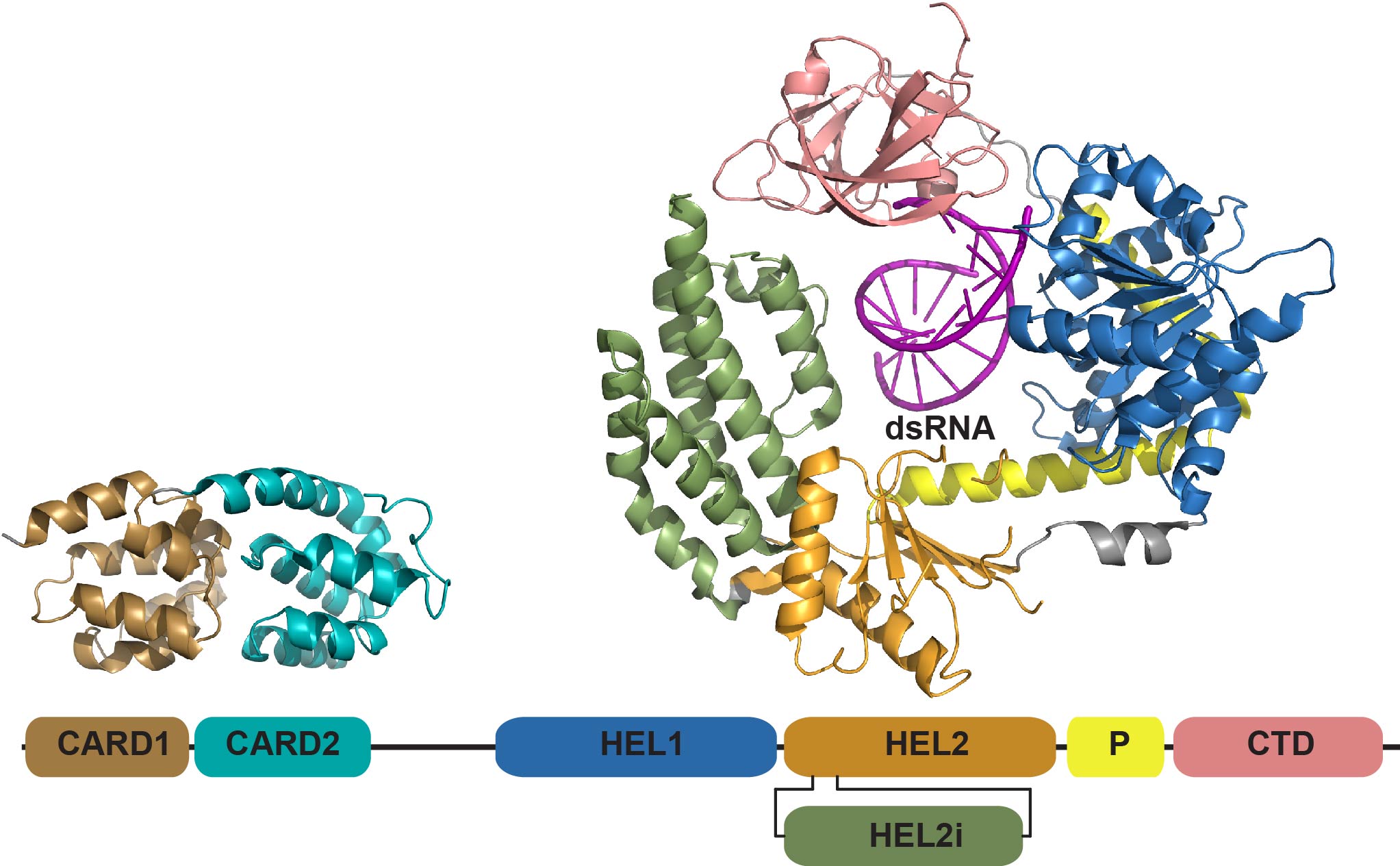
Analytical Ultracentrifugation
Analytical ultracentrifugation (AUC) is a classical technique to define the size, shape, and interactions of biological macromolecules. Recent advances in instrumentation and data analysis algorithms have greatly enhanced the power of AUC to characterize complex macromolecular assemblies. We use instruments in the Biophysics Facility extensively to define the protein-protein and protein-RNA interactions that govern activation of the innate immunity pathway. We also collaborate with other groups to apply advanced AUC methods to diverse research problems.